Rat RAB11A ELISA Kit (RTFI00430)
- SKU:
- RTFI00430
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- P62494
- Sensitivity:
- 0.938ng/ml
- Range:
- 1.563-100ng/ml
- ELISA Type:
- Competitive
- Synonyms:
- Rab11a, RAB11A, Ras-related protein Rab-11A, Rab-11, YL8, RAB11
- Reactivity:
- Rat
- Research Area:
- Cell Cycle
Description
Rat RAB11A ELISA Kit
The Rat Rab11a ELISA Kit is a powerful tool for measuring the levels of Rab11a in rat serum, plasma, and cell culture supernatants. With its high sensitivity and specificity, this kit delivers accurate and consistent results, making it ideal for various research applications.Rab11a is a key protein involved in intracellular trafficking, playing a critical role in regulating vesicle transport and membrane recycling. Dysregulation of Rab11a is associated with various diseases, including cancer, neurological disorders, and infectious diseases, making it a valuable biomarker for understanding disease mechanisms and developing targeted therapies.
Overall, the Rat Rab11a ELISA Kit is a reliable and efficient solution for researchers seeking to investigate the role of Rab11a in physiological and pathological processes in rat models.
| Product Name: | Rat Rab11a (Ras-related protein Rab-11A) ELISA Kit |
| Product Code: | RTFI00430 |
| Size: | 96 Assays |
| Target: | Rat Rab11a |
| Alias: | Rab11a, RAB11A, Ras-related protein Rab-11A, Rab-11, YL8, RAB11 |
| Reactivity: | Rat |
| Detection Method: | Competitive ELISA, Coated with Antibody |
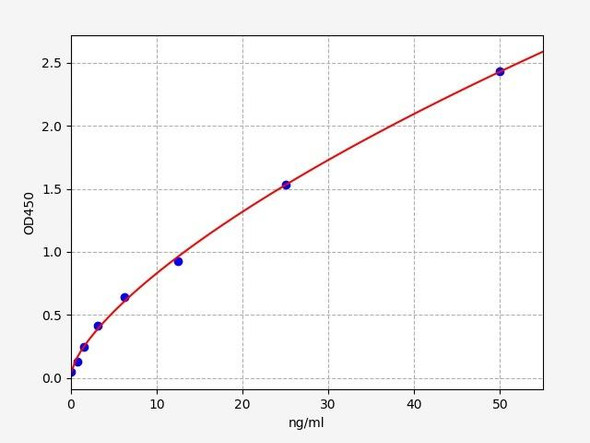
| Sensitivity: | 0.938ng/ml |
| Range: | 1.56-100ng/ml |
| Storage: | 4°C for 6 months |
| Note: | For Research Use Only |
| Recovery: | Matrices listed below were spiked with certain level of Rat Rab11a and the recovery rates were calculated by comparing the measured value to the expected amount of Rat Rab11a in samples. | ||||||||||||||||
| |||||||||||||||||
| Linearity: | The linearity of the kit was assayed by testing samples spiked with appropriate concentration of Rat Rab11a and their serial dilutions. The results were demonstrated by the percentage of calculated concentration to the expected. | ||||||||||||||||
| |||||||||||||||||
| Intra-Assay: | CV <8% | ||||||||||||||||
| Inter-Assay: | CV <10% |
| Uniprot: | P62494 |
| UniProt Protein Function: | RAB11A: Regulates endocytic recycling. May exert its functions by interacting with multiple effector proteins in different complexes. Acts as a major regulator of membrane delivery during cytokinesis. Together with MYO5B and RAB8A participates in epithelial cell polarization. Together with RAB3IP, RAB8A, the exocyst complex, PARD3, PRKCI, ANXA2, CDC42 and DNMBP promotes transcytosis of PODXL to the apical membrane initiation sites (AMIS), apical surface formation and lumenogenesis. Together with MYO5B participates in CFTR trafficking to the plasma membrane and TF (Transferrin) recycling in nonpolarized cells. Required in a complex with MYO5B and RAB11FIP2 for the transport of NPC1L1 to the plasma membrane. Participates in the sorting and basolateral transport of CDH1 from the Golgi apparatus to the plasma membrane. Regulates the recycling of FCGRT (receptor of Fc region of monomeric Ig G) to basolateral membranes. Interacts with RIP11 and STXBP6. Interacts with SGSM1, SGSM2 and SGSM3. Interacts with EXOC6 in a GTP- dependent manner. Interacts with RAB11FIP1, RAB11FIP2, RAB11FIP3 (via its C-terminus) and RAB11FIP4. Interacts with EVI5; EVI5 and RAB11FIP3 may be mutually exclusive and compete for binding RAB11A. Interacts with SPE39//C14orf133. Interacts with MYO5B. Found in a complex with MYO5B and CFTR. Interacts with NPC1L1. Interacts (GDP-bound form) with ZFYVE27. Interacts with BIRC6/bruce. Belongs to the small GTPase superfamily. Rab family. |
| UniProt Protein Details: | Protein type:G protein; G protein, monomeric; G protein, monomeric, Rab; Motility/polarity/chemotaxis Chromosomal Location of Human Ortholog: 8q24 Cellular Component: axon; centrosome; cleavage furrow; cytoplasm; cytoplasmic vesicle; Golgi apparatus; kinetochore microtubule; membrane; mitochondrion; multivesicular body; perinuclear region of cytoplasm; phagocytic vesicle; protein complex; recycling endosome; spindle pole; trans-Golgi network; transport vesicle; vesicle Molecular Function:GTP binding; GTPase activity; microtubule binding; myosin V binding Biological Process: astral microtubule organization and biogenesis; establishment of vesicle localization; exocytosis; melanosome transport; mitotic metaphase plate congression; neurite development; positive regulation of axon extension; regulation of long-term neuronal synaptic plasticity; regulation of protein transport; vesicle-mediated transport |
| UniProt Code: | P62494 |
| NCBI GenInfo Identifier: | 13592148 |
| NCBI Gene ID: | 81830 |
| NCBI Accession: | NP_112414.1 |
| UniProt Secondary Accession: | P62494,P24410, Q9JLX1, |
| Molecular Weight: | 24,394 Da |
| NCBI Full Name: | ras-related protein Rab-11A |
| UniProt Protein Name: | Ras-related protein Rab-11A |
| UniProt Synonym Protein Names: | 24KG |
| Protein Family: | Ras-related protein |
| UniProt Gene Name: | Rab11a |
| Step | Procedure |
| 1. | Set standard, test sample and control (zero) wells on the pre-coated plate respectively, and then, record their positions. It is recommended to measure each standard and sample in duplicate. Wash plate 2 times before adding standard, sample and control (zero) wells! |
| 2. | Add Sample and Biotin-detection antibody: Add 50µL of Standard, Blank or Sample per well. The blank well is added with Sample Dilution Buffer. Immediately add 50 µL of biotin-labelled antibody working solution to each well. Cover with the plate sealer provided. Gently tap the plate to ensure thorough mixing. Incubate for 45 minutes at 37°C. (Solutions are added to the bottom of micro-ELISA plate well, avoid touching plate walls and foaming). |
| 3. | Wash: Aspirate each well and wash, repeating the process three times. Wash by filling each well with Wash Buffer (approximately 350µL) using a squirt bottle, multi-channel pipette, manifold dispenser or automated washer. Complete removal of liquid at each step is essential to good performance. After the last wash, remove any remaining Wash Buffer by aspirating or decanting. Invert the plate and pat it against thick clean absorbent paper. |
| 4. | HRP-Streptavidin Conjugate(SABC): Add 100µL of SABC working solution to each well. Cover with a new Plate sealer. Incubate for 30 minutes at 37°C. |
| 5. | Wash: Repeat the aspiration/wash process for five times. |
| 6. | TMB Substrate: Add 90µL of TMB Substrate to each well. Cover with a new plate sealer. Incubate for about 10-20 minutes at 37°C. Protect from light. The reaction time can be shortened or extended according to the actual color change, but not more than 30 minutes. When apparent gradient appeared in standard wells, you can terminate the reaction. |
| 7. | Stop: Add 50µL of Stop Solution to each well. Color turn to yellow immediately. The adding order of stop solution should be as the same as the substrate solution. |
| 8. | OD Measurement: Determine the optical density (OD Value) of each well at once, using a microplate reader set to 450 nm. You should open the microplate reader ahead, preheat the instrument, and set the testing parameters. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum: | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clotovernight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Removeserum and assay promptly or aliquot and store the samples at-80°C. Avoid multiple freeze-thaw cycles. |
| Plasma: | Collect plasma using EDTA or heparin as an anti-coagulant. Centrifuge samples at 4°C for 15 mins at 1000 - g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles.Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid: | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell Culture Supernatant: | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell Lysates: | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20°C. |
| Tissue Homogenates: | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenizein 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or-80°C. |
| Tissue Lysates: | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk: | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |