Rat PKR / EIF2AK2 ELISA Kit
- SKU:
- RTFI00316
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- Q63184
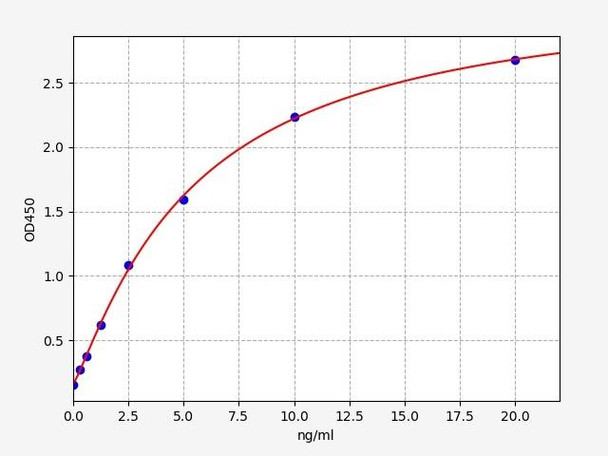
- Sensitivity:
- 0.188ng/ml
- Range:
- 0.313-20ng/ml
- ELISA Type:
- Sandwich
- Synonyms:
- Eif2ak2, PRKR, PRKRA, PKR, EIF2AK1, p68 kinase, double stranded RNA activated protein kinase, eIF-2A protein kinase 2, eukaryotic translation initiation factor 2-alpha kinase 2, P1, eIF-2A protein kinase, interferon-inducible double stranded RNA depe
- Reactivity:
- Rat
- Research Area:
- Immunology
Description
Rat PKR/EIF2AK2 ELISA Kit
The Rat PKR (EIF2AK2) ELISA Kit is specifically designed for the precise measurement of PKR levels in rat serum, plasma, and cell culture supernatants. This kit offers exceptional sensitivity and accuracy, guaranteeing consistent and dependable results for a variety of research purposes.PKR, also known as EIF2AK2, is a vital enzyme that plays a crucial role in the regulation of the immune response to viral infections and cellular stress. Abnormal PKR activity has been linked to various diseases including viral infections, autoimmune disorders, and cancer, making it a valuable biomarker for studying these conditions and exploring potential therapeutic interventions.
With the Rat PKR (EIF2AK2) ELISA Kit, researchers can confidently assess PKR levels in rat samples with precision and reliability, providing valuable insights for advancing scientific knowledge and developing innovative treatments.
| Product Name: | Rat Eif2ak2 (Interferon-induced, double-stranded RNA-activated protein kinase) ELISA Kit |
| Product Code: | RTFI00316 |
| Size: | 96 Assays |
| Target: | Rat Eif2ak2 |
| Alias: | Eif2ak2, PRKR, PRKRA, PKR, EIF2AK1, p68 kinase, double stranded RNA activated protein kinase, eIF-2A protein kinase 2, eukaryotic translation initiation factor 2-alpha kinase 2, P1, eIF-2A protein kinase, interferon-inducible double stranded RNA dependent, interferon-inducible elF2alpha kinase, Interferon-inducible RNA-dependent protein kinase, Protein kinase RNA-activated |
| Reactivity: | Rat |
| Detection Method: | Sandwich ELISA, Double Antibody |
| Sensitivity: | 0.188ng/ml |
| Range: | 0.313-20ng/ml |
| Storage: | 4°C for 6 months |
| Note: | For Research Use Only |
| Recovery: | Matrices listed below were spiked with certain level of Rat Eif2ak2 and the recovery rates were calculated by comparing the measured value to the expected amount of Rat Eif2ak2 in samples. | ||||||||||||||||
| |||||||||||||||||
| Linearity: | The linearity of the kit was assayed by testing samples spiked with appropriate concentration of Rat Eif2ak2 and their serial dilutions. The results were demonstrated by the percentage of calculated concentration to the expected. | ||||||||||||||||
| |||||||||||||||||
| Intra-Assay: | CV <8% | ||||||||||||||||
| Inter-Assay: | CV <10% |
| Uniprot: | Q63184 |
| UniProt Protein Function: | Function: IFN-induced dsRNA-dependent serine/threonine-protein kinase which plays a key role in the innate immune response to viral infection and is also involved in the regulation of signal transduction, apoptosis, cell proliferation and differentiation. Exerts its antiviral activity on a wide range of DNA and RNA viruses including west nile virus (WNV), sindbis virus (SV), foot-and-mouth virus (FMDV), semliki Forest virus (SFV) and lymphocytic choriomeningitis virus (LCMV). Inhibits viral replication via phosphorylation of the alpha subunit of eukaryotic initiation factor 2 (EIF2S1), this phosphorylation impairs the recycling of EIF2S1 between successive rounds of initiation leading to inhibition of translation which eventually results in shutdown of cellular and viral protein synthesis. Also phosphorylates other substrates including p53/TP53, PPP2R5A, DHX9, ILF3 and IRS1. In addition to serine/threonine-protein kinase activity, also has tyrosine-protein kinase activity and phosphorylates CDK1 at 'Tyr-4' upon DNA damage, facilitating its ubiquitination and proteosomal degradation. Either as an adapter protein and/or via its kinase activity, can regulate various signaling pathways (p38 MAP kinase, NF-kappa-B and insulin signaling pathways) and transcription factors (JUN, STAT1, STAT3, IRF1, ATF3) involved in the expression of genes encoding proinflammatory cytokines and IFNs. Activates the NF-kappa-B pathway via interaction with IKBKB and TRAF family of proteins and activates the p38 MAP kinase pathway via interaction with MAP2K6. Can act as both a positive and negative regulator of the insulin signaling pathway (ISP). Negatively regulates ISP by inducing the inhibitory phosphorylation of insulin receptor substrate 1 (IRS1) at 'Ser-312' and positively regulates ISP via phosphorylation of PPP2R5A which activates FOXO1, which in turn up-regulates the expression of insulin receptor substrate 2 (IRS2). Can regulate NLRP3 inflammasome assembly and the activation of NLRP3, NLRP1, AIM2 and NLRC4 inflammasomes. Can trigger apoptosis via FADD-mediated activation of CASP8. Plays a role in the regulation of the cytoskeleton by binding to gelsolin (GSN), sequestering the protein in an inactive conformation away from actin. Regulates proliferation, differentiation and survival of hematopoietic stem/progenitor cells and induction of cytokines and chemokines |
| UniProt Protein Details: | By similarity. Plays a role in cortex-dependent memory consolidation. Ref.3 Catalytic activity: ATP + a protein = ADP + a phosphoprotein.ATP + a [protein]-L-tyrosine = ADP + a [protein]-L-tyrosine phosphate. Enzyme regulation: Initially produced in an inactive form and is activated by binding to viral dsRNA, which causes dimerization and autophosphorylation in the activation loop and stimulation of function. ISGylation can activate it in the absence of viral infection. Can also be activated by heparin, proinflammatory stimuli, growth factors, cytokines, oxidative stress and the cellular protein PRKRA. Activity is markedly stimulated by manganese ions. Activation is blocked by the cellular proteins TARBP2, DUS2L, NPM1, NCK1 and ADAR By similarity. Subunit structure: Homodimer By similarity. Interacts with DNAJC3 By similarity. Interacts with STRBP. Forms a complex with FANCA, FANCC, FANCG and HSP70 By similarity. Interacts with ADAR/ADAR1 By similarity. The inactive form interacts with NCK1 and GSN. Interacts (via the kinase catalytic domain) with STAT3 (via SH2 domain), TRAF2 (C-terminus), TRAF5 (C-terminus) and TRAF6 (C-terminus). Interacts with MAP2K6, IKBKB/IKKB, IRS1, NPM1, TARBP2, NLRP1, NLRP3, NLRC4 and AIM2. Interacts (via DRBM 1 domain) with DUS2L (via DRBM domain). Interacts with DHX9 (via N-terminus) and this interaction is dependent upon activation of the kinase By similarity. Ref.2 Subcellular location: Cytoplasm By similarity. Nucleus By similarity. Cytoplasm perinuclear region By similarity. Induction: By type I interferons By similarity. Post-translational modification: Autophosphorylated on several Ser, Thr and Tyr residues. Autophosphorylation of Thr-411 is dependent on Thr-406 and is stimulated by dsRNA binding and dimerization. Autophosphorylation apparently leads to the activation of the kinase. Tyrosine autophosphorylation is essential for efficient dsRNA-binding, dimerization, and kinase activation By similarity. Sequence similarities: Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. GCN2 subfamily.Contains 2 DRBM (double-stranded RNA-binding) domains.Contains 1 protein kinase domain. |
| NCBI Summary: | double-stranded RNA dependent eukaryotic initiation factor 2-kinase [RGD, Feb 2006] |
| UniProt Code: | Q63184 |
| NCBI GenInfo Identifier: | 9506993 |
| NCBI Gene ID: | 54287 |
| NCBI Accession: | NP_062208.1 |
| UniProt Related Accession: | Q63184 |
| Molecular Weight: | 29.9kDa |
| NCBI Full Name: | interferon-induced, double-stranded RNA-activated protein kinase |
| NCBI Synonym Full Names: | eukaryotic translation initiation factor 2-alpha kinase 2 |
| NCBI Official Symbol: | Eif2ak2 |
| NCBI Official Synonym Symbols: | Pkr; Prkr |
| NCBI Protein Information: | interferon-induced, double-stranded RNA-activated protein kinase; eIF-2A protein kinase 2; protein kinase RNA-activated; tyrosine-protein kinase EIF2AK2; interferon-inducible RNA-dependent protein kinase; Protein kinase, interferon-inducible double stranded RNA dependent |
| UniProt Protein Name: | Interferon-induced, double-stranded RNA-activated protein kinase |
| UniProt Synonym Protein Names: | Eukaryotic translation initiation factor 2-alpha kinase 2; eIF-2A protein kinase 2; Interferon-inducible RNA-dependent protein kinase; Protein kinase RNA-activated; PKR; Tyrosine-protein kinase EIF2AK2 |
| UniProt Gene Name: | Eif2ak2 |
| UniProt Entry Name: | E2AK2_RAT |
| Step | Procedure |
| 1. | Set standard, test sample and control (zero) wells on the pre-coated plate respectively, and then, record their positions. It is recommended to measure each standard and sample in duplicate. Wash plate 2 times before adding standard, sample and control (zero) wells! |
| 2. | Aliquot 0.1ml standard solutions into the standard wells. |
| 3. | Add 0.1 ml of Sample / Standard dilution buffer into the control (zero) well. |
| 4. | Add 0.1 ml of properly diluted sample ( Human serum, plasma, tissue homogenates and other biological fluids.) into test sample wells. |
| 5. | Seal the plate with a cover and incubate at 37°C for 90 min. |
| 6. | Remove the cover and discard the plate content, clap the plate on the absorbent filter papers or other absorbent material. Do NOT let the wells completely dry at any time. Wash plate X2. |
| 7. | Add 0.1 ml of Biotin- detection antibody working solution into the above wells (standard, test sample & zero wells). Add the solution at the bottom of each well without touching the side wall. |
| 8. | Seal the plate with a cover and incubate at 37°C for 60 min. |
| 9. | Remove the cover, and wash plate 3 times with Wash buffer. Let wash buffer rest in wells for 1 min between each wash. |
| 10. | Add 0.1 ml of SABC working solution into each well, cover the plate and incubate at 37°C for 30 min. |
| 11. | Remove the cover and wash plate 5 times with Wash buffer, and each time let the wash buffer stay in the wells for 1-2 min. |
| 12. | Add 90 µL of TMB substrate into each well, cover the plate and incubate at 37°C in dark within 10-20 min. (Note: This incubation time is for reference use only, the optimal time should be determined by end user.) And the shades of blue can be seen in the first 3-4 wells (with most concentrated standard solutions), the other wells show no obvious color. |
| 13. | Add 50 µL of Stop solution into each well and mix thoroughly. The color changes into yellow immediately. |
| 14. | Read the O.D. absorbance at 450 nm in a microplate reader immediately after adding the stop solution. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum: | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clotovernight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Removeserum and assay promptly or aliquot and store the samples at-80°C. Avoid multiple freeze-thaw cycles. |
| Plasma: | Collect plasma using EDTA or heparin as an anti-coagulant. Centrifuge samples at 4°C for 15 mins at 1000 × g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles.Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid: | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell Culture Supernatant: | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell Lysates: | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20°C. |
| Tissue Homogenates: | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenizein 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or-80°C. |
| Tissue Lysates: | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk: | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |