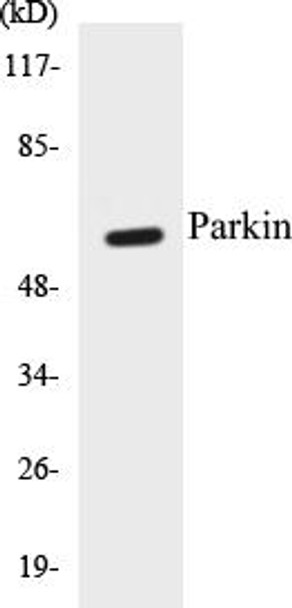
Parkin Colorimetric Cell-Based ELISA Kit
- SKU:
- CBCAB00345
- Product Type:
- ELISA Kit
- ELISA Type:
- Cell Based
- Research Area:
- Autophagy
- Reactivity:
- Human
- Detection Method:
- Colorimetric
Description
Parkin Colorimetric Cell-Based ELISA Kit
The Parkin Colorimetric Cell-Based ELISA Kit is a cutting-edge tool for the precise quantification of parkin levels in cell lysates and tissues. This kit offers exceptional sensitivity and specificity, guaranteeing accurate and consistent results for a variety of research applications.Parkin is a key protein involved in the regulation of mitochondrial quality control and the removal of damaged organelles through the process of mitophagy. Dysregulation of parkin has been linked to various neurodegenerative diseases, including Parkinson's disease, making it a critical biomarker for investigating these conditions and exploring potential therapeutic interventions.
With its advanced technology and user-friendly protocol, the Parkin Colorimetric Cell-Based ELISA Kit is an indispensable asset for researchers seeking to deepen their understanding of parkin biology and its implications for human health. Elevate your research endeavors with this innovative assay kit from AssayGenie.
| Product Name: | Parkin Colorimetric Cell-Based ELISA |
| Product Code: | CBCAB00345 |
| ELISA Type: | Cell-Based |
| Target: | Parkin |
| Reactivity: | Human |
| Dynamic Range: | > 5000 Cells |
| Detection Method: | Colorimetric 450 nmStorage/Stability:4°C/6 Months |
| Format: | 96-Well Microplate |
The Parkin Colorimetric Cell-Based ELISA Kit is a convenient, lysate-free, high throughput and sensitive assay kit that can detect Parkin protein expression profile in cells. The kit can be used for measuring the relative amounts of Parkin in cultured cells as well as screening for the effects that various treatments, inhibitors (ie siRNA or chemicals), or activators have on Parkin.
Qualitative determination of Parkin concentration is achieved by an indirect ELISA format. In essence, Parkin is captured by Parkin-specific primary antibodies while the HRP-conjugated secondary antibodies bind the Fc region of the primary antibody. Through this binding, the HRP enzyme conjugated to the secondary antibody can catalyze a colorimetric reaction upon substrate addition. Due to the qualitative nature of the Cell-Based ELISA, multiple normalization methods are needed:
| 1. | A monoclonal antibody specific for human GAPDH is included to serve as an internal positive control in normalizing the target absorbance values. |
| 2. | Following the colorimetric measurement of HRP activity via substrate addition, the Crystal Violet whole-cell staining method may be used to determine cell density. After staining, the results can be analysed by normalizing the absorbance values to cell amounts, by which the plating difference can be adjusted. |
| Database Information: | Gene ID: 5071, UniProt ID: O60260, OMIM: 602544, Unigene: Hs.132954 |
| Gene Symbol: | PRKN2 |
| Sub Type: | None |
| UniProt Protein Function: | PARK2: a component of a multiprotein E3 ubiquitin ligase complex, catalyzing the covalent attachment of ubiquitin moieties onto substrate proteins, such as BCL2, SYT11, CCNE1, GPR37, STUB1, a 22 kDa O-linked glycosylated isoform of SNCAIP, SEPT5, ZNF746 and AIMP2. Mediates monoubiquitination as well as 'Lys-48'-linked and 'Lys-63'-linked polyubiquitination of substrates depending on the context. Participates in the removal and/or detoxification of abnormally folded or damaged protein by mediating 'Lys-63'-linked polyubiquitination of misfolded proteins such as PARK7: 'Lys-63'- linked polyubiquitinated misfolded proteins are then recognized by HDAC6, leading to their recruitment to aggresomes, followed by degradation. Mediates 'Lys-63'-linked polyubiquitination of SNCAIP, possibly playing a role in Lewy-body formation. Mediates monoubiquitination of BCL2, thereby acting as a positive regulator of autophagy. Promotes the autophagic degradation of dysfunctional depolarized mitochondria. Mediates 'Lys-48'-linked polyubiquitination of ZNF746, followed by degradation of ZNF746 by the proteasome; possibly playing a role in role in regulation of neuron death. Limits the production of reactive oxygen species (ROS). Loss of this ubiquitin ligase activity appears to be the mechanism underlying pathogenesis of PARK2. May protect neurons against alpha synuclein toxicity, proteasomal dysfunction, GPR37 accumulation, and kainate-induced excitotoxicity. May play a role in controlling neurotransmitter trafficking at the presynaptic terminal and in calcium-dependent exocytosis. Regulates cyclin-E during neuronal apoptosis. May represent a tumor suppressor gene. Forms an E3 ubiquitin ligase complex with UBE2L3 or UBE2L6. Mediates 'Lys-63'-linked polyubiquitination by associating with UBE2V1. Part of a SCF-like complex, consisting of PARK2, CUL1 and FBXW7. Part of a complex, including STUB1, HSP70 and GPR37. The amount of STUB1 in the complex increases during ER stress. STUB1 promotes the dissociation of HSP70 from PARK2 and GPR37, thus facilitating PARK2-mediated GPR37 ubiquitination. HSP70 transiently associates with unfolded GPR37 and inhibits the E3 activity of PARK2, whereas, STUB1 enhances the E3 activity of PARK2 through promotion of dissociation of HSP70 from PARK2-GPR37 complexes. Interacts with PSMD4 and PACRG. Interacts with LRRK2. Interacts with RANBP2. Interacts with SUMO1 but not SUMO2, which promotes nuclear localization and autoubiquitination. Interacts (via first RING- type domain) with AIMP2 (via N-terminus). Interacts with PSMA7 and RNF41. Interacts with PINK1. Highly expressed in the brain including the substantia nigra. Expressed in heart, testis and skeletal muscle. Expression is down-regulated or absent in tumor biopsies, and absent in the brain of PARK2 patients. Overexpression protects dopamine neurons from kainate-mediated apoptosis. Found in serum. Belongs to the RBR family. Parkin subfamily. 6 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:Ubiquitin ligase; Ligase; Ubiquitin conjugating system; EC 6.3.2.19; EC 6.3.2.- Chromosomal Location of Human Ortholog: 6q25.2-q27 Cellular Component: Golgi apparatus; neuron projection; mitochondrion; perinuclear region of cytoplasm; endoplasmic reticulum; cytoplasm; SCF ubiquitin ligase complex; nucleus; cytosol; ubiquitin ligase complex Molecular Function:tubulin binding; identical protein binding; ubiquitin binding; zinc ion binding; histone deacetylase binding; ubiquitin-protein ligase activity; Hsp70 protein binding; actin binding; protein kinase binding; PDZ domain binding; protein binding; G-protein-coupled receptor binding; ubiquitin conjugating enzyme binding; chaperone binding; ubiquitin protein ligase binding; heat shock protein binding; kinase binding; SH3 domain binding; ligase activity Biological Process: protein monoubiquitination; proteasomal ubiquitin-dependent protein catabolic process; negative regulation of JNK cascade; negative regulation of actin filament bundle formation; startle response; central nervous system development; protein polyubiquitination; adult locomotory behavior; regulation of protein ubiquitination; protein ubiquitination during ubiquitin-dependent protein catabolic process; regulation of neurotransmitter secretion; protein ubiquitination; mitochondrion localization; norepinephrine metabolic process; dopamine metabolic process; regulation of dopamine secretion; negative regulation of insulin secretion; negative regulation of glucokinase activity; zinc ion homeostasis; negative regulation of protein amino acid phosphorylation; regulation of lipid transport; dopamine uptake; negative regulation of neuron apoptosis; positive regulation of DNA binding; mitochondrion organization and biogenesis; mitochondrial fission; synaptic transmission, glutamatergic; protein autoubiquitination; positive regulation of I-kappaB kinase/NF-kappaB cascade; transcription, DNA-dependent; protein stabilization; learning; regulation of protein transport; cellular protein catabolic process; positive regulation of proteasomal ubiquitin-dependent protein catabolic process; mitochondrion degradation; positive regulation of transcription from RNA polymerase II promoter; regulation of autophagy; response to oxidative stress Disease: Parkinson Disease 2, Autosomal Recessive Juvenile; Leprosy, Susceptibility To, 2; Lung Cancer; Ovarian Cancer |
| NCBI Summary: | The precise function of this gene is unknown; however, the encoded protein is a component of a multiprotein E3 ubiquitin ligase complex that mediates the targeting of substrate proteins for proteasomal degradation. Mutations in this gene are known to cause Parkinson disease and autosomal recessive juvenile Parkinson disease. Alternative splicing of this gene produces multiple transcript variants encoding distinct isoforms. Additional splice variants of this gene have been described but currently lack transcript support. [provided by RefSeq, Jul 2008] |
| UniProt Code: | O60260 |
| NCBI GenInfo Identifier: | 116242725 |
| NCBI Gene ID: | 5071 |
| NCBI Accession: | O60260.2 |
| UniProt Secondary Accession: | O60260,Q5TFV8, Q5VVX4, Q6Q2I6, Q8NI41, Q8NI43, Q8NI44 Q8WW07, A3FG77, A8K975, D3JZW7, D3K2X0, |
| UniProt Related Accession: | O60260 |
| Molecular Weight: | 465 |
| NCBI Full Name: | E3 ubiquitin-protein ligase parkin |
| NCBI Synonym Full Names: | parkin RBR E3 ubiquitin protein ligase |
| NCBI Official Symbol: | PARK2 |
| NCBI Official Synonym Symbols: | PDJ; PRKN; AR-JP; LPRS2 |
| NCBI Protein Information: | E3 ubiquitin-protein ligase parkin; parkinson juvenile disease protein 2; parkinson protein 2, E3 ubiquitin protein ligase (parkin); Parkinson disease (autosomal recessive, juvenile) 2, parkin |
| UniProt Protein Name: | E3 ubiquitin-protein ligase parkin |
| UniProt Synonym Protein Names: | Parkinson juvenile disease protein 2; Parkinson disease protein 2 |
| Protein Family: | E3 ubiquitin-protein ligase |
| UniProt Gene Name: | PARK2 |
| UniProt Entry Name: | PRKN2_HUMAN |
| Component | Quantity |
| 96-Well Cell Culture Clear-Bottom Microplate | 2 plates |
| 10X TBS | 24 mL |
| Quenching Buffer | 24 mL |
| Blocking Buffer | 50 mL |
| 15X Wash Buffer | 50 mL |
| Primary Antibody Diluent | 12 mL |
| 100x Anti-Phospho Target Antibody | 60 µL |
| 100x Anti-Target Antibody | 60 µL |
| Anti-GAPDH Antibody | 60 µL |
| HRP-Conjugated Anti-Rabbit IgG Antibody | 12 mL |
| HRP-Conjugated Anti-Mouse IgG Antibody | 12 mL |
| SDS Solution | 12 mL |
| Stop Solution | 24 mL |
| Ready-to-Use Substrate | 12 mL |
| Crystal Violet Solution | 12 mL |
| Adhesive Plate Seals | 2 seals |
The following materials and/or equipment are NOT provided in this kit but are necessary to successfully conduct the experiment:
- Microplate reader able to measure absorbance at 450 nm and/or 595 nm for Crystal Violet Cell Staining (Optional)
- Micropipettes with capability of measuring volumes ranging from 1 µL to 1 ml
- 37% formaldehyde (Sigma Cat# F-8775) or formaldehyde from other sources
- Squirt bottle, manifold dispenser, multichannel pipette reservoir or automated microplate washer
- Graph paper or computer software capable of generating or displaying logarithmic functions
- Absorbent papers or vacuum aspirator
- Test tubes or microfuge tubes capable of storing ≥1 ml
- Poly-L-Lysine (Sigma Cat# P4832 for suspension cells)
- Orbital shaker (optional)
- Deionized or sterile water
*Note: Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
| Step | Procedure |
| 1. | Seed 200 µL of 20,000 adherent cells in culture medium in each well of a 96-well plate. The plates included in the kit are sterile and treated for cell culture. For suspension cells and loosely attached cells, coat the plates with 100 µL of 10 µg/ml Poly-L-Lysine (not included) to each well of a 96-well plate for 30 minutes at 37°C prior to adding cells. |
| 2. | Incubate the cells for overnight at 37°C, 5% CO2. |
| 3. | Treat the cells as desired. |
| 4. | Remove the cell culture medium and rinse with 200 µL of 1x TBS, twice. |
| 5. | Fix the cells by incubating with 100 µL of Fixing Solution for 20 minutes at room temperature. The 4% formaldehyde is used for adherent cells and 8% formaldehyde is used for suspension cells and loosely attached cells. |
| 6. | Remove the Fixing Solution and wash the plate 3 times with 200 µL 1x Wash Buffer for five minutes each time with gentle shaking on the orbital shaker. The plate can be stored at 4°C for a week. |
| 7. | Add 100 µL of Quenching Buffer and incubate for 20 minutes at room temperature. |
| 8. | Wash the plate 3 times with 1x Wash Buffer for 5 minutes each time. |
| 9. | Add 200 µL of Blocking Buffer and incubate for 1 hour at room temperature. |
| 10. | Wash 3 times with 200 µL of 1x Wash Buffer for 5 minutes each time. |
| 11. | Add 50 µL of 1x primary antibodies (Anti-Parkin Antibody and/or Anti-GAPDH Antibody) to the corresponding wells, cover with Parafilm and incubate for 16 hours (overnight) at 4°C. If the target expression is known to be high, incubate for 2 hours at room temperature. |
| 12. | Wash 3 times with 200 µL of 1x Wash Buffer for 5 minutes each time. |
| 13. | Add 50 µL of 1x secondary antibodies (HRP-Conjugated AntiRabbit IgG Antibody or HRP-Conjugated Anti-Mouse IgG Antibody) to corresponding wells and incubate for 1.5 hours at room temperature. |
| 14. | Wash 3 times with 200 µL of 1x Wash Buffer for 5 minutes each time. |
| 15. | Add 50 µL of Ready-to-Use Substrate to each well and incubate for 30 minutes at room temperature in the dark. |
| 16. | Add 50 µL of Stop Solution to each well and read OD at 450 nm immediately using the microplate reader. |
(Additional Crystal Violet staining may be performed if desired – details of this may be found in the kit technical manual.)