Description
Mouse Insulin receptor (Insr) ELISA Kit
The Mouse Insulin Receptor (INSR) ELISA Kit is a powerful tool for measuring insulin receptor levels in mouse serum, plasma, and cell culture supernatants. This kit provides accurate and reliable results with high sensitivity and specificity, making it ideal for various research applications.The insulin receptor is a key protein involved in regulating glucose metabolism and is essential for maintaining proper blood sugar levels. Dysregulation of the insulin receptor is linked to conditions such as diabetes, obesity, and metabolic syndrome, making it a valuable biomarker for studying these conditions and developing potential treatments.
With its easy-to-use format and robust performance, the Mouse INSR ELISA Kit is a valuable asset for researchers studying insulin signaling pathways and investigating potential therapeutic interventions for metabolic disorders.
| Product Name: | Mouse Insulin receptor (Insr) ELISA Kit |
| SKU: | MOEB0657 |
| Size: | 96T |
| Target: | Mouse Insulin receptor (Insr) |
| Synonyms: | CD220, IR |
| Assay Type: | Sandwich |
| Detection Method: | ELISA |
| Reactivity: | Mouse |
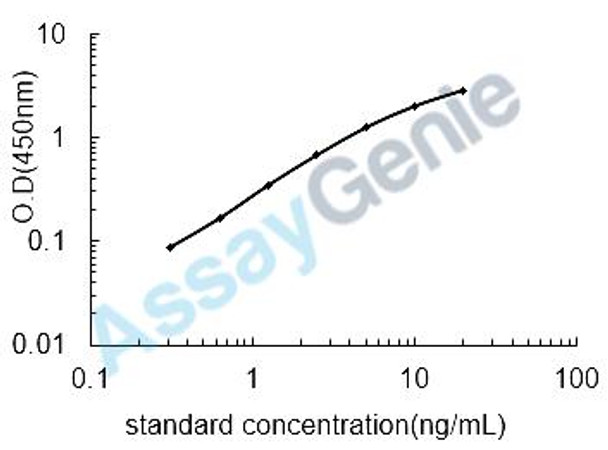
| Detection Range: | 0.312-20ng/mL |
| Sensitivity: | 0.158ng/mL |
| Intra CV: | 4.2% | ||||||||||||||||||||
| Inter CV: | 7.4% | ||||||||||||||||||||
| Linearity: |
| ||||||||||||||||||||
| Recovery: |
| ||||||||||||||||||||
| Function: | Receptor tyrosine kinase which mediates the pleiotropic actions of insulin. Binding of insulin leads to phosphorylation of several intracellular substrates, including, insulin receptor substrates (IRS1, 2, 3, 4), SHC, GAB1, CBL and other signaling intermediates. Each of these phosphorylated proteins serve as docking proteins for other signaling proteins that contain Src-homology-2 domains (SH2 domain) that specifically recognize different phosphotyrosine residues, including the p85 regulatory subunit of PI3K and SHP2. Phosphorylation of IRSs proteins lead to the activation of two main signaling pathways: the PI3K-AKT/PKB pathway, which is responsible for most of the metabolic actions of insulin, and the Ras-MAPK pathway, which regulates expression of some genes and cooperates with the PI3K pathway to control cell growth and differentiation. Binding of the SH2 domains of PI3K to phosphotyrosines on IRS1 leads to the activation of PI3K and the generation of phosphatidylinositol-(3, 4, 5)-triphosphate (PIP3), a lipid second messenger, which activates several PIP3-dependent serine/threonine kinases, such as PDPK1 and subsequently AKT/PKB. The net effect of this pathway is to produce a translocation of the glucose transporter SLC2A4/GLUT4 from cytoplasmic vesicles to the cell membrane to facilitate glucose transport. Moreover, upon insulin stimulation, activated AKT/PKB is responsible for: anti-apoptotic effect of insulin by inducing phosphorylation of BAD; regulates the expression of gluconeogenic and lipogenic enzymes by controlling the activity of the winged helix or forkhead (FOX) class of transcription factors. Another pathway regulated by PI3K-AKT/PKB activation is mTORC1 signaling pathway which regulates cell growth and metabolism and integrates signals from insulin. AKT mediates insulin-stimulated protein synthesis by phosphorylating TSC2 thereby activating mTORC1 pathway. The Ras/RAF/MAP2K/MAPK pathway is mainly involved in mediating cell growth, survival and cellular differentiation of insulin. Phosphorylated IRS1 recruits GRB2/SOS complex, which triggers the activation of the Ras/RAF/MAP2K/MAPK pathway. In addition to binding insulin, the insulin receptor can bind insulin-like growth factors (IGFI and IGFII). When present in a hybrid receptor with IGF1R, binds IGF1. |
| Uniprot: | P15208 |
| Sample Type: | Serum, plasma, tissue homogenates, cell culture supernates and other biological fluids |
| Specificity: | Natural and recombinant mouse Insulin receptor |
| Sub Unit: | Tetramer of 2 alpha and 2 beta chains linked by disulfide bonds. The alpha chains carry the insulin-binding regions, while the beta chains carry the kinase domain. Forms a hybrid receptor with IGF1R, the hybrid is a tetramer consisting of 1 alpha chain and 1 beta chain of INSR and 1 alpha chain and 1 beta chain of IGF1R. Interacts with SORBS1 but dissociates from it following insulin stimulation. Binds SH2B2 (By similarity). Activated form of INSR interacts (via Tyr-989) with the PTB/PID domains of IRS1 and SHC1. The sequences surrounding the phosphorylated NPXY motif contribute differentially to either IRS1 or SHC1 recognition. Interacts (via tyrosines in the C-terminus) with IRS2 (via PTB domain and 591-786 AA); the 591-786 would be the primary anchor of IRS2 to INSR while the PTB domain would have a stabilizing action on the interaction with INSR. Interacts with the SH2 domains of the 85 kDa regulatory subunit of PI3K (PIK3R1) in vitro, when autophosphorylated on tyrosine residues. Interacts with SOCS7 (By similarity). Interacts (via the phosphorylated Tyr-989), with SOCS3. Interacts (via the phosphorylated Tyr-1175, Tyr-1179, Tyr-1180) with SOCS1. Interacts with CAV2 (tyrosine-phosphorylated form); the interaction is increased with 'Tyr-27'phosphorylation of CAV2 (By similarity). Interacts with ARRB2. Interacts with GRB10; this interaction blocks the association between IRS1/IRS2 and INSR, significantly reduces insulin-stimulated tyrosine phosphorylation of IRS1 and IRS2 and thus decreases insulin signaling (By similarity). Interacts with GRB7 (By similarity). Interacts with PDPK1 (By similarity). Interacts (via Tyr-1180) with GRB14 (via BPS domain); this interaction protects the tyrosines in the activation loop from dephosphorylation, but promotes dephosphorylation of Tyr-989, this results in decreased interaction with, and phosphorylation of, IRS1 (By similarity). Interacts (via subunit alpha) with ENPP1 (via 485-599 AA); this interaction blocks autophosphorylation (By similarity). Interacts with PTPRE; this interaction is dependent of Tyr-1175, Tyr-1179 and Tyr-1180 of the INSR (By similarity). Interacts with STAT5B (via SH2 domain) (By similarity). Interacts with PTPRF. |
| Research Area: | Cardiovascular |
| Subcellular Location: | Cell membrane Single-pass type I membrane protein |
| Storage: | Please see kit components below for exact storage details |
| Note: | For research use only |
| UniProt Protein Function: | INSR: a receptor tyrosine kinase that mediates the pleiotropic actions of insulin. Binding of insulin leads to phosphorylation of several intracellular substrates, including, insulin receptor substrates (IRS1, 2, 3, 4), SHC, GAB1, CBL and other signaling intermediates. Each of these phosphorylated proteins serve as docking proteins for other signaling proteins that contain Src-homology-2 domains (SH2 domain) that specifically recognize different phosphotyrosines residues, including the p85 regulatory subunit of PI3K and SHP2. Phosphorylation of IRSs proteins lead to the activation of two main signaling pathways: the PI3K-AKT pathway, which is responsible for most of the metabolic actions of insulin, and the Ras-MAPK pathway, which regulates expression of some genes and cooperates with the PI3K pathway to control cell growth and differentiation. In addition to binding insulin, the insulin receptor can bind insulin-like growth factors (IGFI and IGFII). The holoenzyme is cleaved into two chains, the alpha and beta subunits. The active complex is a tetramer containing 2 alpha and 2 beta chains linked by disulfide bonds. The alpha chains constitute the ligand- binding domain, while the beta chains carry the kinase domain. Interacts with SORBS1 but dissociates from it following insulin stimulation. Familial mutations associated with insulin resistant diabetes, acanthosis nigricans, pineal hyperplasia, and polycystic ovary syndrome. SNP variants may be associated with polycystic ovary syndrome, atypical migraine and diabetic hyperlipidemia. Mutations also cause leprechaunism, a severe insulin resistance syndrome causing growth retardation and death in early infancy. Two isoforms of the human protein are produced by alternative splicing. The Short isoform has a higher affinity for insulin than the longer. Isoform Long and isoform Short are predominantly expressed in tissue targets of insulin metabolic effects: liver, adipose tissue and skeletal muscle but are also expressed in the peripheral nerve, kidney, pulmonary alveoli, pancreatic acini, placenta vascular endothelium, fibroblasts, monocytes, granulocytes, erythrocytes and skin. Isoform Short is preferentially expressed in fetal cells such as fetal fibroblasts, muscle, liver and kidney. Found as a hybrid receptor with IGF1R in muscle, heart, kidney, adipose tissue, skeletal muscle, hepatoma, fibroblasts, spleen and placenta. Overexpressed in several tumors, including breast, colon, lung, ovary, and thyroid carcinomas.Protein type: Kinase, protein; Protein kinase, TK; Membrane protein, integral; EC 2.7.10.1; Protein kinase, tyrosine (receptor); TK group; InsR familyCellular Component: caveola; cell soma; cytosol; endosome; integral to plasma membrane; intracellular membrane-bound organelle; membrane; nucleus; plasma membrane; receptor complex; synapseMolecular Function: 3-phosphoinositide-dependent protein kinase binding; ATP binding; GTP binding; insulin binding; insulin receptor activity; insulin receptor substrate binding; insulin-like growth factor I binding; insulin-like growth factor II binding; insulin-like growth factor receptor binding; lipoic acid binding; phosphoinositide 3-kinase binding; protein binding; protein complex binding; protein domain specific binding; protein kinase activity; protein kinase binding; protein phosphatase binding; protein-tyrosine kinase activity; PTB domain binding; receptor signaling protein tyrosine kinase activityBiological Process: activation of MAPK activity; activation of protein kinase activity; activation of protein kinase B; adrenal gland development; cellular response to insulin stimulus; epidermis development; exocrine pancreas development; G-protein coupled receptor protein signaling pathway; glucose homeostasis; heart morphogenesis; insulin receptor signaling pathway; male gonad development; male sex determination; negative regulation of protein amino acid phosphorylation; negative regulation of transporter activity; organ morphogenesis; peptidyl-tyrosine phosphorylation; positive regulation of cell migration; positive regulation of cell proliferation; positive regulation of developmental growth; positive regulation of DNA replication; positive regulation of glucose import; positive regulation of glycogen biosynthetic process; positive regulation of glycolysis; positive regulation of MAPKKK cascade; positive regulation of meiotic cell cycle; positive regulation of mitosis; positive regulation of nitric oxide biosynthetic process; positive regulation of phosphorylation; positive regulation of protein amino acid phosphorylation; positive regulation of protein kinase B signaling cascade; positive regulation of transcription, DNA-dependent; protein amino acid autophosphorylation; protein amino acid phosphorylation; protein heterotetramerization; regulation of embryonic development; regulation of hydrogen peroxide metabolic process; regulation of transcription, DNA-dependent; transformation of host cell by virus |
| UniProt Protein Details: | |
| NCBI Summary: | |
| UniProt Code: | P15208 |
| NCBI GenInfo Identifier: | 408360149 |
| NCBI Gene ID: | 16337 |
| NCBI Accession: | P15208.2 |
| UniProt Secondary Accession: | P15208,F8VPU4 |
| UniProt Related Accession: | P15208 |
| Molecular Weight: | 155,610 Da |
| NCBI Full Name: | Insulin receptor |
| NCBI Synonym Full Names: | insulin receptor |
| NCBI Official Symbol: | Insr |
| NCBI Official Synonym Symbols: | IR; IR-A; IR-B; CD220; 4932439J01Rik; D630014A15Rik |
| NCBI Protein Information: | insulin receptor |
| UniProt Protein Name: | Insulin receptor |
| UniProt Synonym Protein Names: | CD_antigen: CD220 |
| Protein Family: | Insulin receptor |
| UniProt Gene Name: | Insr |
| UniProt Entry Name: | INSR_MOUSE |
| Component | Quantity (96 Assays) | Storage |
| ELISA Microplate (Dismountable) | 8×12 strips | -20°C |
| Lyophilized Standard | 2 | -20°C |
| Sample Diluent | 20ml | -20°C |
| Assay Diluent A | 10mL | -20°C |
| Assay Diluent B | 10mL | -20°C |
| Detection Reagent A | 120µL | -20°C |
| Detection Reagent B | 120µL | -20°C |
| Wash Buffer | 30mL | 4°C |
| Substrate | 10mL | 4°C |
| Stop Solution | 10mL | 4°C |
| Plate Sealer | 5 | - |
Other materials and equipment required:
- Microplate reader with 450 nm wavelength filter
- Multichannel Pipette, Pipette, microcentrifuge tubes and disposable pipette tips
- Incubator
- Deionized or distilled water
- Absorbent paper
- Buffer resevoir
*Note: The below protocol is a sample protocol. Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
Allow all reagents to reach room temperature (Please do not dissolve the reagents at 37°C directly). All the reagents should be mixed thoroughly by gently swirling before pipetting. Avoid foaming. Keep appropriate numbers of strips for 1 experiment and remove extra strips from microtiter plate. Removed strips should be resealed and stored at -20°C until the kits expiry date. Prepare all reagents, working standards and samples as directed in the previous sections. Please predict the concentration before assaying. If values for these are not within the range of the standard curve, users must determine the optimal sample dilutions for their experiments. We recommend running all samples in duplicate.
| Step | |
| 1. | Add Sample: Add 100µL of Standard, Blank, or Sample per well. The blank well is added with Sample diluent. Solutions are added to the bottom of micro ELISA plate well, avoid inside wall touching and foaming as possible. Mix it gently. Cover the plate with sealer we provided. Incubate for 120 minutes at 37°C. |
| 2. | Remove the liquid from each well, don't wash. Add 100µL of Detection Reagent A working solution to each well. Cover with the Plate sealer. Gently tap the plate to ensure thorough mixing. Incubate for 1 hour at 37°C. Note: if Detection Reagent A appears cloudy warm to room temperature until solution is uniform. |
| 3. | Aspirate each well and wash, repeating the process three times. Wash by filling each well with Wash Buffer (approximately 400µL) (a squirt bottle, multi-channel pipette,manifold dispenser or automated washer are needed). Complete removal of liquid at each step is essential. After the last wash, completely remove remaining Wash Buffer by aspirating or decanting. Invert the plate and pat it against thick clean absorbent paper. |
| 4. | Add 100µL of Detection Reagent B working solution to each well. Cover with the Plate sealer. Incubate for 60 minutes at 37°C. |
| 5. | Repeat the wash process for five times as conducted in step 3. |
| 6. | Add 90µL of Substrate Solution to each well. Cover with a new Plate sealer and incubate for 10-20 minutes at 37°C. Protect the plate from light. The reaction time can be shortened or extended according to the actual color change, but this should not exceed more than 30 minutes. When apparent gradient appears in standard wells, user should terminatethe reaction. |
| 7. | Add 50µL of Stop Solution to each well. If color change does not appear uniform, gently tap the plate to ensure thorough mixing. |
| 8. | Determine the optical density (OD value) of each well at once, using a micro-plate reader set to 450 nm. User should open the micro-plate reader in advance, preheat the instrument, and set the testing parameters. |
| 9. | After experiment, store all reagents according to the specified storage temperature respectively until their expiry. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clot overnight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Remove serum and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. |
| Plasma | Collect plasma using EDTA or heparin as an anticoagulant. Centrifuge samples at 4°C for 15 mins at 1000 × g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell culture supernatant | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell lysates | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Tissue homogenates | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenize in 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or -80°C. |
| Tissue lysates | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |