Human X-box-binding protein 1 (XBP1) ELISA Kit (HUEB2618)
- SKU:
- HUEB2618
- Product Type:
- ELISA Kit
- Size:
- 96 Assays
- Uniprot:
- P17861
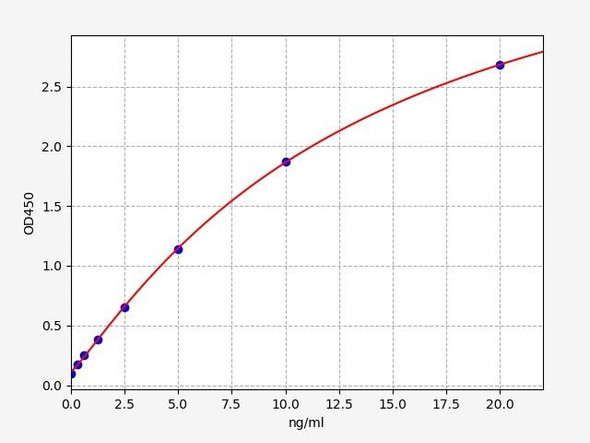
- Range:
- 0.156-10 ng/mL
- ELISA Type:
- Sandwich
- Synonyms:
- XBP1, HTF, TREB5, XBP2, TREB5, X-box binding protein 1, X-box-binding protein 1, XBP-1, XBP2Tax-responsive element-binding protein 5
- Reactivity:
- Human
Description
Human X-box-binding protein 1 (XBP1) ELISA Kit
The Human X-box Binding Protein 1 (XBP1) ELISA Kit is specifically designed for the precise measurement of XBP1 levels in human serum, plasma, and cell culture supernatants. This kit offers exceptional sensitivity and specificity, ensuring consistent and accurate results for a variety of research applications.XBP1 is a key transcription factor involved in the unfolded protein response, playing a crucial role in regulating cellular stress and maintaining protein homeostasis.
Dysregulation of XBP1 has been implicated in various diseases, including cancer, neurodegenerative disorders, and autoimmune conditions, highlighting its significance as a potential biomarker for disease progression and therapeutic development.Overall, the Human XBP1 ELISA Kit provides researchers with a valuable tool for investigating the role of XBP1 in health and disease, offering reliable data to support further studies and advancements in biomedical research.
| Product Name: | Human X-box-binding protein 1 (XBP1) ELISA Kit |
| SKU: | HUEB2618 |
| Size: | 96T |
| Target: | Human X-box-binding protein 1 (XBP1) |
| Synonyms: | Tax-responsive element-binding protein 5, TREB-5, XBP-1, TREB5, XBP2 |
| Assay Type: | Sandwich |
| Detection Method: | ELISA |
| Reactivity: | Human |
| Detection Range: | 0.156-10ng/mL |
| Sensitivity: | 0.078ng/mL |
| Intra CV: | 5.5% | ||||||||||||||||||||
| Inter CV: | 7.9% | ||||||||||||||||||||
| Linearity: |
| ||||||||||||||||||||
| Recovery: |
| ||||||||||||||||||||
| Function: | Isoform 2: functions as a stress-inducible potent transcriptional activator during endoplasmic reticulum (ER) stress by inducing unfolded protein response (UPR) target genes via binding to the UPR element (UPRE). Up-regulates target genes encoding ER chaperones and ER-associated degradation (ERAD) components to enhance the capacity of productive folding and degradation mechanism, respectively, in order to maintain the homeostasis of the ER under ER stress (PubMed:11779464, PubMed:25239945). Plays a role in the production of immunoglobulins and interleukin-6 in the presence of stimuli required for plasma cell differentiation (By similarity). Induces phospholipid biosynthesis and ER expansion (PubMed:15466483). Contributes to the VEGF-induced endothelial cell (EC) growth and proliferation in a Akt/GSK-dependent and/or -independent signaling pathway, respectively, leading to beta-catenin nuclear translocation and E2F2 gene expression (PubMed:23529610). Promotes umbilical vein EC apoptosis and atherosclerotisis development in a caspase-dependent signaling pathway, and contributes to VEGF-induced EC proliferation and angiogenesis in adult tissues under ischemic conditions (PubMed:19416856, PubMed:23529610). Involved in the regulation of endostatin-induced autophagy in EC through BECN1 transcriptional activation (PubMed:23184933). Plays a role as an oncogene by promoting tumor progression: stimulates zinc finger protein SNAI1 transcription to induce epithelial-to-mesenchymal (EMT) transition, cell migration and invasion of breast cancer cells (PubMed:25280941). Involved in adipocyte differentiation by regulating lipogenic gene expression during lactation. Plays a role in the survival of both dopaminergic neurons of the substantia nigra pars compacta (SNpc), by maintaining protein homeostasis and of myeloma cells. Increases insulin sensitivity in the liver as a response to a high carbohydrate diet, resulting in improved glucose tolerance. Improves also glucose homeostasis in an ER stress- and/or insulin-independent manner through both binding and proteasome-induced degradation of the transcription factor FOXO1, hence resulting in suppression of gluconeogenic genes expression and in a reduction of blood glucose levels. Controls the induction of de novo fatty acid synthesis in hepatocytes by regulating the expression of a subset of lipogenic genes in an ER stress- and UPR-independent manner (By similarity). Associates preferentially to the HDAC3 gene promoter region in a disturbed flow-dependent manner (PubMed:25190803). Binds to the BECN1 gene promoter region (PubMed:23184933). Binds to the CDH5/VE-cadherin gene promoter region (PubMed:19416856). Binds to the ER stress response element (ERSE) upon ER stress (PubMed:11779464). Binds to the 5'-CCACG-3' motif in the PPARG promoter. |
| Uniprot: | P17861 |
| Sample Type: | Serum, plasma, tissue homogenates, cell culture supernates and other biological fluids |
| Specificity: | Natural and recombinant human X-box-binding protein 1 |
| Sub Unit: | Isoform 2 interacts with SIRT1. Isoform 2 interacts with PIK3R1 and PIK3R2; the interactions are direct and induce translocation of XBP1 isoform 2 into the nucleus and the unfolded protein response (UPR) XBP1-dependent target genes activation in a ER stress- and/or insulin-dependent but PI3K-independent manner. Isoform 2 interacts with FOXO1; the interaction is direct and leads to FOXO1 ubiquitination and degradation via the proteasome pathway in hepatocytes (By similarity). Isoform 1 interacts with HM13 (PubMed:25239945). Isoform 1 interacts with RNF139; the interaction induces ubiquitination and degradation of isoform 1 (PubMed:25239945). Isoform 1 interacts (via luminal domain) with DERL1; the interaction obviates the need for ectodomain shedding prior HM13/SPP-mediated XBP1 isoform 1 cleavage (PubMed:25239945). Isoform 1 interacts with isoform 2; the interaction sequesters isoform 2 from the nucleus and enhances isoform 2 degradation in the cytoplasm (PubMed:16461360, PubMed:25239945). Isoform 1 interacts with HDAC3 and AKT1; the interactions occur in endothelial cell (EC) under disturbed flow (PubMed:25190803). Isoform 1 interacts with the oncoprotein FOS (PubMed:1903538). Isoform 2 interacts with ATF6; the interaction occurs in a ER stress-dependent manner and is required for DNA binding to the unfolded protein response element (UPRE) (PubMed:17765680). Isoform 2 interacts with PIK3R1; the interaction is direct and induces translocation of XBP1 isoform 2 into the nucleus and the unfolded protein response (UPR) XBP1-dependent target genes activation in a ER stress- and/or insulin-dependent but PI3K-independent manner (PubMed:20348923). |
| Research Area: | Epigenetics |
| Subcellular Location: | X-box-binding protein 1, cytoplasmic form Cytoplasm Nucleus Localizes in the cytoplasm and nucleus after HM13/SPP-mediated intramembranaire proteolytic cleavage of isoform 1 (PubMed:25239945). |
| Storage: | Please see kit components below for exact storage details |
| Note: | For research use only |
| UniProt Protein Function: | XBP1: a transcription factor essential for hepatocyte growth, the differentiation of plasma cells, immunoglobulin secretion, and the unfolded protein response (UPR). XBP1 mRNA is spliced by IRE1 during the UPR to generate a new C-terminus, converting it into a potent unfolded-protein response transcriptional activator and triggering growth arrest and apoptosis. Only the spliced form of XBP1 can activate the UPR efficiently. Activates UPR target genes via direct binding to the UPR element (UPRE). Binds DNA preferably to the CRE-like element 5'-GATGACGTG[TG]N(3)[AT]T-3', and also to some TPA response elements (TRE). Binds to the HLA DR-alpha promoter. Binds to the Tax-responsive element (TRE) of HTLV-I. Up-regulated by ATF6 via direct binding to the ERSE in response to endoplasmic reticulum stress. Genetic variations in XBP1 could be associated with susceptibility to major affective disorder type 7 (MAFD7). Major affective disorders represent a class of mental disorders characterized by a disturbance in mood as their predominant feature. Two human isoforms are produced by alternative splicing. Isoform 1 is also known as XBP-1U. Isoform 2, also known as XBP-1S, is produced by IRE1 in response to endoplasmic reticulum stress. IRE1 cleaves a 26-bp fragment causing a frameshift of the mRNA transcript. |
| UniProt Protein Details: | Protein type:DNA-binding; Transcription factor Chromosomal Location of Human Ortholog: 22q12.1|22q12 Cellular Component: nucleoplasm; endoplasmic reticulum membrane; endoplasmic reticulum; cytoplasm; integral to membrane; integral to endoplasmic reticulum membrane; nucleus; cytosol Molecular Function:protein binding; protein homodimerization activity; protease binding; DNA binding; protein heterodimerization activity; chromatin DNA binding; ubiquitin protein ligase binding; estrogen receptor binding; transcription factor activity; protein kinase binding Biological Process: ubiquitin-dependent protein catabolic process; transcription from RNA polymerase II promoter; phosphoinositide 3-kinase cascade; muscle development; apoptosis; exocrine pancreas development; positive regulation of transcription of target genes involved in unfolded protein response; regulation of protein stability; negative regulation of transcription from RNA polymerase II promoter; cellular response to glucose starvation; protein transport; serotonin secretion, neurotransmission; positive regulation of MHC class II biosynthetic process; positive regulation of transcription factor import into nucleus; angiogenesis; response to electrical stimulus; cell growth; positive regulation of autophagy; fatty acid biosynthetic process; positive regulation of TOR signaling pathway; response to drug; positive regulation of histone methylation; protein destabilization; unfolded protein response; organelle organization and biogenesis; cellular response to nutrient; positive regulation of immunoglobulin secretion; liver development; positive regulation of immunoglobulin production; cholesterol homeostasis; unfolded protein response, activation of signaling protein activity; cellular protein metabolic process; cellular response to insulin stimulus; positive regulation of T cell differentiation; fatty acid homeostasis; endothelial cell proliferation; positive regulation of B cell differentiation; positive regulation of fat cell differentiation; neuron development; autophagy; immune response; positive regulation of transcription from RNA polymerase II promoter; positive regulation of protein amino acid phosphorylation; sterol homeostasis; vascular endothelial growth factor receptor signaling pathway; negative regulation of apoptosis Disease: Major Affective Disorder 7 |
| NCBI Summary: | This gene encodes a transcription factor that regulates MHC class II genes by binding to a promoter element referred to as an X box. This gene product is a bZIP protein, which was also identified as a cellular transcription factor that binds to an enhancer in the promoter of the T cell leukemia virus type 1 promoter. It may increase expression of viral proteins by acting as the DNA binding partner of a viral transactivator. It has been found that upon accumulation of unfolded proteins in the endoplasmic reticulum (ER), the mRNA of this gene is processed to an active form by an unconventional splicing mechanism that is mediated by the endonuclease inositol-requiring enzyme 1 (IRE1). The resulting loss of 26 nt from the spliced mRNA causes a frame-shift and an isoform XBP1(S), which is the functionally active transcription factor. The isoform encoded by the unspliced mRNA, XBP1(U), is constitutively expressed, and thought to function as a negative feedback regulator of XBP1(S), which shuts off transcription of target genes during the recovery phase of ER stress. A pseudogene of XBP1 has been identified and localized to chromosome 5. [provided by RefSeq, Jul 2008] |
| UniProt Code: | P17861 |
| NCBI GenInfo Identifier: | 60416406 |
| NCBI Gene ID: | 7494 |
| NCBI Accession: | P17861.2 |
| UniProt Secondary Accession: | P17861,Q8WYK6, Q969P1, Q96BD7, |
| UniProt Related Accession: | P17861 |
| Molecular Weight: | 40,148 Da |
| NCBI Full Name: | X-box-binding protein 1 |
| NCBI Synonym Full Names: | X-box binding protein 1 |
| NCBI Official Symbol: | XBP1 |
| NCBI Official Synonym Symbols: | XBP2; TREB5; XBP-1 |
| NCBI Protein Information: | X-box-binding protein 1; tax-responsive element-binding protein 5 |
| UniProt Protein Name: | X-box-binding protein 1 |
| UniProt Synonym Protein Names: | Tax-responsive element-binding protein 5 |
| Protein Family: | X-box-binding protein |
| UniProt Gene Name: | XBP1 |
| UniProt Entry Name: | XBP1_HUMAN |
| Component | Quantity (96 Assays) | Storage |
| ELISA Microplate (Dismountable) | 8×12 strips | -20°C |
| Lyophilized Standard | 2 | -20°C |
| Sample Diluent | 20ml | -20°C |
| Assay Diluent A | 10mL | -20°C |
| Assay Diluent B | 10mL | -20°C |
| Detection Reagent A | 120µL | -20°C |
| Detection Reagent B | 120µL | -20°C |
| Wash Buffer | 30mL | 4°C |
| Substrate | 10mL | 4°C |
| Stop Solution | 10mL | 4°C |
| Plate Sealer | 5 | - |
Other materials and equipment required:
- Microplate reader with 450 nm wavelength filter
- Multichannel Pipette, Pipette, microcentrifuge tubes and disposable pipette tips
- Incubator
- Deionized or distilled water
- Absorbent paper
- Buffer resevoir
*Note: The below protocol is a sample protocol. Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
Allow all reagents to reach room temperature (Please do not dissolve the reagents at 37°C directly). All the reagents should be mixed thoroughly by gently swirling before pipetting. Avoid foaming. Keep appropriate numbers of strips for 1 experiment and remove extra strips from microtiter plate. Removed strips should be resealed and stored at -20°C until the kits expiry date. Prepare all reagents, working standards and samples as directed in the previous sections. Please predict the concentration before assaying. If values for these are not within the range of the standard curve, users must determine the optimal sample dilutions for their experiments. We recommend running all samples in duplicate.
| Step | |
| 1. | Add Sample: Add 100µL of Standard, Blank, or Sample per well. The blank well is added with Sample diluent. Solutions are added to the bottom of micro ELISA plate well, avoid inside wall touching and foaming as possible. Mix it gently. Cover the plate with sealer we provided. Incubate for 120 minutes at 37°C. |
| 2. | Remove the liquid from each well, don't wash. Add 100µL of Detection Reagent A working solution to each well. Cover with the Plate sealer. Gently tap the plate to ensure thorough mixing. Incubate for 1 hour at 37°C. Note: if Detection Reagent A appears cloudy warm to room temperature until solution is uniform. |
| 3. | Aspirate each well and wash, repeating the process three times. Wash by filling each well with Wash Buffer (approximately 400µL) (a squirt bottle, multi-channel pipette,manifold dispenser or automated washer are needed). Complete removal of liquid at each step is essential. After the last wash, completely remove remaining Wash Buffer by aspirating or decanting. Invert the plate and pat it against thick clean absorbent paper. |
| 4. | Add 100µL of Detection Reagent B working solution to each well. Cover with the Plate sealer. Incubate for 60 minutes at 37°C. |
| 5. | Repeat the wash process for five times as conducted in step 3. |
| 6. | Add 90µL of Substrate Solution to each well. Cover with a new Plate sealer and incubate for 10-20 minutes at 37°C. Protect the plate from light. The reaction time can be shortened or extended according to the actual color change, but this should not exceed more than 30 minutes. When apparent gradient appears in standard wells, user should terminatethe reaction. |
| 7. | Add 50µL of Stop Solution to each well. If color change does not appear uniform, gently tap the plate to ensure thorough mixing. |
| 8. | Determine the optical density (OD value) of each well at once, using a micro-plate reader set to 450 nm. User should open the micro-plate reader in advance, preheat the instrument, and set the testing parameters. |
| 9. | After experiment, store all reagents according to the specified storage temperature respectively until their expiry. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clot overnight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Remove serum and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. |
| Plasma | Collect plasma using EDTA or heparin as an anticoagulant. Centrifuge samples at 4°C for 15 mins at 1000 × g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell culture supernatant | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell lysates | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Tissue homogenates | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenize in 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or -80°C. |
| Tissue lysates | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |
| ELISA |
| Human XBP1 ELISA Kit |