Description
Human Interferon-induced helicase C domain-containing protein 1 (IFIH1) ELISA Kit
The Human Interferon Induced Helicase C domain-containing protein 1 (IFIH1) ELISA Kit is a reliable tool for measuring levels of IFIH1 in human serum, plasma, and cell culture supernatants. This kit offers high sensitivity and specificity, ensuring accurate and reproducible results for a variety of research applications.IFIH1 is a key protein involved in the innate immune response to viral infections, particularly in the detection of viral RNA. Dysregulation of IFIH1 has been linked to autoimmune diseases such as type 1 diabetes and systemic lupus erythematosus.
By accurately measuring IFIH1 levels, researchers can better understand its role in immune responses and autoimmune diseases, potentially leading to the development of new diagnostic tools and therapeutic interventions. The Human IFIH1 ELISA Kit is a valuable asset for researchers studying viral infections and autoimmune disorders.
| Product Name: | Human Interferon-induced helicase C domain-containing protein 1 (IFIH1) ELISA Kit |
| SKU: | HUEB2376 |
| Size: | 96T |
| Target: | Human Interferon-induced helicase C domain-containing protein 1 (IFIH1) |
| Synonyms: | Clinically amyopathic dermatomyositis autoantigen 140 kDa, Helicase with 2 CARD domains, Interferon-induced with helicase C domain protein 1, Melanoma differentiation-associated protein 5, Murabutide down-regulated protein, RIG-I-like receptor 2, RNA helicase-DEAD box protein 116, CADM-140 autoantigen, Helicard, MDA-5, RLR-2, MDA5, RH116 |
| Assay Type: | Sandwich |
| Detection Method: | ELISA |
| Reactivity: | Human |
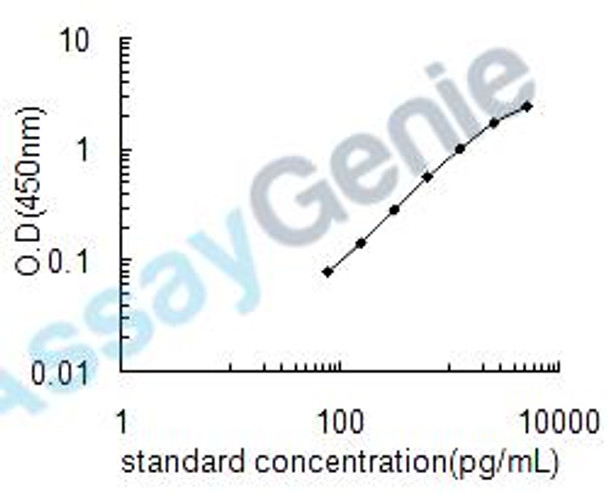
| Detection Range: | 78-5000pg/mL |
| Sensitivity: | 39pg/mL |
| Intra CV: | 4.5% | ||||||||||||||||||||
| Inter CV: | 7.7% | ||||||||||||||||||||
| Linearity: |
| ||||||||||||||||||||
| Recovery: |
| ||||||||||||||||||||
| Function: | Innate immune receptor which acts as a cytoplasmic sensor of viral nucleic acids and plays a major role in sensing viral infection and in the activation of a cascade of antiviral responses including the induction of type I interferons and proinflammatory cytokines. Its ligands include mRNA lacking 2'-O-methylation at their 5' cap and long-dsRNA (>1 kb in length). Upon ligand binding it associates with mitochondria antiviral signaling protein (MAVS/IPS1) which activates the IKK-related kinases: TBK1 and IKBKE which phosphorylate interferon regulatory factors: IRF3 and IRF7 which in turn activate transcription of antiviral immunological genes, including interferons (IFNs); IFN-alpha and IFN-beta. Responsible for detecting the Picornaviridae family members such as encephalomyocarditis virus (EMCV) and mengo encephalomyocarditis virus (ENMG). Can also detect other viruses such as dengue virus (DENV), west Nile virus (WNV), and reovirus. Also involved in antiviral signaling in response to viruses containing a dsDNA genome, such as vaccinia virus. Plays an important role in amplifying innate immune signaling through recognition of RNA metabolites that are produced during virus infection by ribonuclease L (RNase L). May play an important role in enhancing natural killer cell function and may be involved in growth inhibition and apoptosis in several tumor cell lines. |
| Uniprot: | Q9BYX4 |
| Sample Type: | Serum, plasma, tissue homogenates, cell culture supernates and other biological fluids |
| Specificity: | Natural and recombinant human Interferon-induced helicase C domain-containing protein 1 |
| Sub Unit: | Monomer in the absence of ligands and homodimerizes in the presence of dsRNA ligands. Can assemble into helical or linear polymeric filaments on long dsRNA. Interacts with MAVS/IPS1. Interacts (via the CARD domains) with TKFC, the interaction is inhibited by viral infection (PubMed:17600090). Interacts with PCBP2. Interacts with NLRC5. Interacts with PIAS2-beta. Interacts with DDX60. Interacts with ANKRD17. Interacts with IKBKE (PubMed:17600090). Interacts with V protein of Simian virus 5, Human parainfluenza virus 2, Mumps virus, Sendai virus and Hendra virus. Binding to paramyxoviruses V proteins prevents IFN-beta induction, and the further establishment of an antiviral state. Interacts with herpes simplex virus 1 protein US11; this interaction prevents the interaction of MAVS/IPS1 to IFIH1. |
| Research Area: | Immunology |
| Subcellular Location: | Cytoplasm Nucleus May be found in the nucleus, during apoptosis. |
| Storage: | Please see kit components below for exact storage details |
| Note: | For research use only |
| UniProt Protein Function: | IFIH1: Innate immune receptor which acts as a cytoplasmic sensor of viral nucleic acids and plays a major role in sensing viral infection and in the activation of a cascade of antiviral responses including the induction of type I interferons and proinflammatory cytokines. Its ligands include mRNA lacking 2'-O- methylation at their 5' cap and long-dsRNA (>1 kb in length). Upon ligand binding it associates with mitochondria antiviral signaling protein (MAVS/IPS1) which activates the IKK-related kinases: TBK1 and IKBKE which phosphorylate interferon regulatory factors: IRF3 and IRF7 which in turn activate transcription of antiviral immunological genes, including interferons (IFNs); IFN-alpha and IFN-beta. Responsible for detecting the Picornaviridae family members such as encephalomyocarditis virus (EMCV) and mengo encephalomyocarditis virus (ENMG). Can also detect other viruses such as dengue virus (DENV), west Nile virus (WNV), and reovirus. Also involved in antiviral signaling in response to viruses containing a dsDNA genome, such as vaccinia virus. Plays an important role in amplifying innate immune signaling through recognition of RNA metabolites that are produced during virus infection by ribonuclease L (RNase L). May play an important role in enhancing natural killer cell function and may be involved in growth inhibition and apoptosis in several tumor cell lines. Monomer in the absence of ligands and homodimerizes in the presence of dsRNA ligands. Can assemble into helical or linear polymeric filaments on long dsRNA. Interacts with MAVS/IPS1. Interacts with V protein of Simian virus 5, Human parainfluenza virus 2, Mumps virus, Sendai virus and Hendra virus. Binding to paramyxoviruses V proteins prevents IFN-beta induction, and the further establishment of an antiviral state. Interacts with PCBP2. Interacts with NLRC5. Interacts with PIAS2-beta. Interacts with DDX60. By interferon (IFN) and TNF. Widely expressed, at a low level. Expression is detected at slightly highest levels in placenta, pancreas and spleen and at barely levels in detectable brain, testis and lung. Belongs to the helicase family. RLR subfamily. 2 isoforms of the human protein are produced by alternative splicing. |
| UniProt Protein Details: | Protein type:EC 3.6.4.13; Helicase; Apoptosis Chromosomal Location of Human Ortholog: 2q24 Cellular Component: cytosol; nucleus Molecular Function:ATP binding; DNA binding; double-stranded RNA binding; helicase activity; protein binding; ribonucleoprotein binding; single-stranded RNA binding; zinc ion binding Biological Process: detection of virus; innate immune response; negative regulation of interferon type I production; positive regulation of interferon-alpha production; positive regulation of interferon-beta production; protein sumoylation; response to virus; viral reproduction Disease: Aicardi-goutieres Syndrome 7; Singleton-merten Syndrome 1 |
| NCBI Summary: | DEAD box proteins, characterized by the conserved motif Asp-Glu-Ala-Asp (DEAD), are putative RNA helicases. They are implicated in a number of cellular processes involving alteration of RNA secondary structure such as translation initiation, nuclear and mitochondrial splicing, and ribosome and spliceosome assembly. Based on their distribution patterns, some members of this family are believed to be involved in embryogenesis, spermatogenesis, and cellular growth and division. This gene encodes a DEAD box protein that is upregulated in response to treatment with beta-interferon and a protein kinase C-activating compound, mezerein. Irreversible reprogramming of melanomas can be achieved by treatment with both these agents; treatment with either agent alone only achieves reversible differentiation. Genetic variation in this gene is associated with diabetes mellitus insulin-dependent type 19. [provided by RefSeq, Jul 2012] |
| UniProt Code: | Q9BYX4 |
| NCBI GenInfo Identifier: | 134047802 |
| NCBI Gene ID: | 64135 |
| NCBI Accession: | Q9BYX4.3 |
| UniProt Secondary Accession: | Q9BYX4,Q2NKL6, Q6DC96, Q86X56, Q96MX8, Q9H3G6, |
| UniProt Related Accession: | Q9BYX4 |
| Molecular Weight: | 25,129 Da |
| NCBI Full Name: | Interferon-induced helicase C domain-containing protein 1 |
| NCBI Synonym Full Names: | interferon induced, with helicase C domain 1 |
| NCBI Official Symbol: | IFIH1 |
| NCBI Official Synonym Symbols: | AGS7; Hlcd; MDA5; MDA-5; RLR-2; IDDM19; SGMRT1 |
| NCBI Protein Information: | interferon-induced helicase C domain-containing protein 1 |
| UniProt Protein Name: | Interferon-induced helicase C domain-containing protein 1 |
| UniProt Synonym Protein Names: | Clinically amyopathic dermatomyositis autoantigen 140 kDa; CADM-140 autoantigen; Helicase with 2 CARD domains; Helicard; Interferon-induced with helicase C domain protein 1; Melanoma differentiation-associated protein 5; MDA-5; Murabutide down-regulated protein; RIG-I-like receptor 2; RLR-2; RNA helicase-DEAD box protein 116 |
| UniProt Gene Name: | IFIH1 |
| UniProt Entry Name: | IFIH1_HUMAN |
| Component | Quantity (96 Assays) | Storage |
| ELISA Microplate (Dismountable) | 8×12 strips | -20°C |
| Lyophilized Standard | 2 | -20°C |
| Sample Diluent | 20ml | -20°C |
| Assay Diluent A | 10mL | -20°C |
| Assay Diluent B | 10mL | -20°C |
| Detection Reagent A | 120µL | -20°C |
| Detection Reagent B | 120µL | -20°C |
| Wash Buffer | 30mL | 4°C |
| Substrate | 10mL | 4°C |
| Stop Solution | 10mL | 4°C |
| Plate Sealer | 5 | - |
Other materials and equipment required:
- Microplate reader with 450 nm wavelength filter
- Multichannel Pipette, Pipette, microcentrifuge tubes and disposable pipette tips
- Incubator
- Deionized or distilled water
- Absorbent paper
- Buffer resevoir
*Note: The below protocol is a sample protocol. Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
Allow all reagents to reach room temperature (Please do not dissolve the reagents at 37°C directly). All the reagents should be mixed thoroughly by gently swirling before pipetting. Avoid foaming. Keep appropriate numbers of strips for 1 experiment and remove extra strips from microtiter plate. Removed strips should be resealed and stored at -20°C until the kits expiry date. Prepare all reagents, working standards and samples as directed in the previous sections. Please predict the concentration before assaying. If values for these are not within the range of the standard curve, users must determine the optimal sample dilutions for their experiments. We recommend running all samples in duplicate.
| Step | |
| 1. | Add Sample: Add 100µL of Standard, Blank, or Sample per well. The blank well is added with Sample diluent. Solutions are added to the bottom of micro ELISA plate well, avoid inside wall touching and foaming as possible. Mix it gently. Cover the plate with sealer we provided. Incubate for 120 minutes at 37°C. |
| 2. | Remove the liquid from each well, don't wash. Add 100µL of Detection Reagent A working solution to each well. Cover with the Plate sealer. Gently tap the plate to ensure thorough mixing. Incubate for 1 hour at 37°C. Note: if Detection Reagent A appears cloudy warm to room temperature until solution is uniform. |
| 3. | Aspirate each well and wash, repeating the process three times. Wash by filling each well with Wash Buffer (approximately 400µL) (a squirt bottle, multi-channel pipette,manifold dispenser or automated washer are needed). Complete removal of liquid at each step is essential. After the last wash, completely remove remaining Wash Buffer by aspirating or decanting. Invert the plate and pat it against thick clean absorbent paper. |
| 4. | Add 100µL of Detection Reagent B working solution to each well. Cover with the Plate sealer. Incubate for 60 minutes at 37°C. |
| 5. | Repeat the wash process for five times as conducted in step 3. |
| 6. | Add 90µL of Substrate Solution to each well. Cover with a new Plate sealer and incubate for 10-20 minutes at 37°C. Protect the plate from light. The reaction time can be shortened or extended according to the actual color change, but this should not exceed more than 30 minutes. When apparent gradient appears in standard wells, user should terminatethe reaction. |
| 7. | Add 50µL of Stop Solution to each well. If color change does not appear uniform, gently tap the plate to ensure thorough mixing. |
| 8. | Determine the optical density (OD value) of each well at once, using a micro-plate reader set to 450 nm. User should open the micro-plate reader in advance, preheat the instrument, and set the testing parameters. |
| 9. | After experiment, store all reagents according to the specified storage temperature respectively until their expiry. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clot overnight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Remove serum and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. |
| Plasma | Collect plasma using EDTA or heparin as an anticoagulant. Centrifuge samples at 4°C for 15 mins at 1000 × g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell culture supernatant | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell lysates | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Tissue homogenates | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenize in 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or -80°C. |
| Tissue lysates | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |