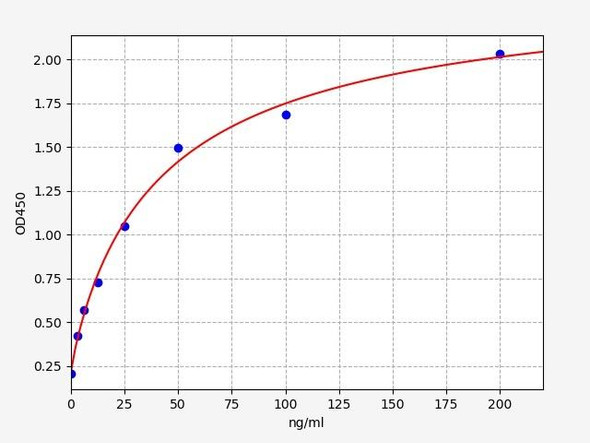
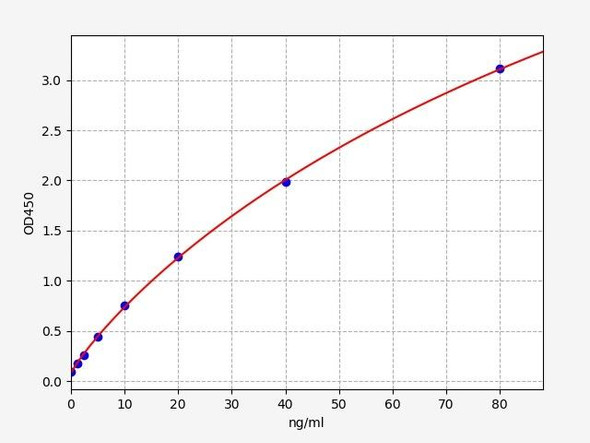
Description
Human Apolipoprotein E (APOE) ELISA Kit
The Human Apolipoprotein E (ApoE) ELISA Kit is a powerful tool for quantitative detection of ApoE levels in human serum, plasma, and cell culture supernatants. With its high sensitivity and specificity, this kit provides accurate and reliable results for a variety of research applications.Apolipoprotein E is a key player in lipid metabolism and is associated with various diseases, including cardiovascular disease and Alzheimer's disease.
By measuring ApoE levels, researchers can gain valuable insights into the role of this protein in disease pathogenesis and potentially identify new therapeutic targets.Overall, the Human Apolipoprotein E (ApoE) ELISA Kit offers a convenient and efficient solution for studying ApoE biology and its implications in health and disease.
| Product Name: | Human Apolipoprotein E (APOE) ELISA Kit |
| SKU: | HUEB0208 |
| Size: | 96T |
| Target: | Human Apolipoprotein E (APOE) |
| Synonyms: | Apo-E |
| Assay Type: | Sandwich |
| Detection Method: | ELISA |
| Reactivity: | Human |
| Detection Range: | 7.8-500ng/mL |
| Sensitivity: | 3.1ng/mL |
| Intra CV: | 4.1% | ||||||||||||||||||||
| Inter CV: | 7.8% | ||||||||||||||||||||
| Linearity: |
| ||||||||||||||||||||
| Recovery: |
| ||||||||||||||||||||
| Function: | Mediates the binding, internalization, and catabolism of lipoprotein particles. It can serve as a ligand for the LDL (apo B/E) receptor and for the specific apo-E receptor (chylomicron remnant) of hepatic tissues. |
| Uniprot: | P02649 |
| Sample Type: | Serum, plasma, tissue homogenates, cell culture supernates and other biological fluids |
| Specificity: | Natural and recombinant human Apolipoprotein E |
| Research Area: | Cardiovascular |
| Subcellular Location: | Secreted |
| Storage: | Please see kit components below for exact storage details |
| Note: | For research use only |
| UniProt Protein Function: | Function: Mediates the binding, internalization, and catabolism of lipoprotein particles. It can serve as a ligand for the LDL (apo B/E) receptor and for the specific apo-E receptor (chylomicron remnant) of hepatic tissues. |
| UniProt Protein Details: | Subcellular location: Secreted. Tissue specificity: Occurs in all lipoprotein fractions in plasma. It constitutes 10-20% of very low density lipoproteins (VLDL) and 1-2% of high density lipoproteins (HDL). APOE is produced in most organs. Significant quantities are produced in liver, brain, spleen, lung, adrenal, ovary, kidney and muscle. Ref.18 Post-translational modification: Synthesized with the sialic acid attached by O-glycosidic linkage and is subsequently desialylated in plasma. O-glycosylated with core 1 or possibly core 8 glycans. Thr-307 is a minor glycosylation site compared to Ser-308. Ref.19Glycated in plasma VLDL of normal subjects, and of hyperglycemic diabetic patients at a higher level (2-3 fold).Phosphorylation sites are present in the extracelllular medium. Polymorphism: Three common APOE alleles have been identified: APOE*2, APOE*3, and APOE*4. The corresponding three major isoforms, E2, E3, and E4, are recognized according to their relative position after isoelectric focusing. Different mutations causing the same migration pattern after isoelectric focusing define different isoform subtypes. The most common isoform is E3 and is present in 40-90% of the population. Common APOE variants influence lipoprotein metabolism in healthy individuals. Involvement in Disease: Defects in APOE are a cause of hyperlipoproteinemia type 3 (HLPP3) [ MIM:107741]; also known as familial dysbetalipoproteinemia. Individuals with HLPP3 are clinically characterized by xanthomas, yellowish lipid deposits in the palmar crease, or less specific on tendons and on elbows. The disorder rarely manifests before the third decade in men. In women, it is usually expressed only after the menopause. The vast majority of the patients are homozygous for APOE*2 alleles. More severe cases of HLPP3 have also been observed in individuals heterozygous for rare APOE variants. The influence of APOE on lipid levels is often suggested to have major implications for the risk of coronary artery disease (CAD). Individuals carrying the common APOE*4 variant are at higher risk of CAD. Ref.16 Ref.26 Ref.27 Ref.29Genetic variations in APOE are associated with Alzheimer disease type 2 (AD2) [ MIM:104310]. It is a late-onset neurodegenerative disorder characterized by progressive dementia, loss of cognitive abilities, and deposition of fibrillar amyloid proteins as intraneuronal neurofibrillary tangles, extracellular amyloid plaques and vascular amyloid deposits. The major constituent of these plaques is the neurotoxic amyloid-beta-APP 40-42 peptide (s), derived proteolytically from the transmembrane precursor protein APP by sequential secretase processing. The cytotoxic C-terminal fragments (CTFs) and the caspase-cleaved products such as C31 derived from APP, are also implicated in neuronal death. Note=The APOE*4 allele is genetically associated with the common late onset familial and sporadic forms of Alzheimer disease. Risk for AD increased from 20% to 90% and mean age at onset decreased from 84 to 68 years with increasing number of APOE*4 alleles in 42 families with late onset AD. Thus APOE*4 gene dose is a major risk factor for late onset AD and, in these families, homozygosity for APOE*4 was virtually sufficient to cause AD by age 80. The mechanism by which APOE*4 participates in pathogenesis is not known. Ref.16Defects in APOE are a cause of sea-blue histiocyte disease (SBHD) [ MIM:269600]; also known as sea-blue histiocytosis. This disorder is characterized by splenomegaly, mild thrombocytopenia and, in the bone marrow, numerous histiocytes containing cytoplasmic granules which stain bright blue with the usual hematologic stains. The syndrome is the consequence of an inherited metabolic defect analogous to Gaucher disease and other sphingolipidoses. Ref.16 Ref.33 Ref.36Defects in APOE are a cause of lipoprotein glomerulopathy (LPG) [ MIM:611771]. LPG is an uncommon kidney disease characterized by proteinuria, progressive kidney failure, and distinctive lipoprotein thrombi in glomerular capillaries. It mainly affects people of Japanese and Chinese origin. The disorder has rarely been described in Caucasians. Ref.16 Ref.30 Ref.32 Ref.37 Sequence similarities: Belongs to the apolipoprotein A1/A4/E family. |
| NCBI Summary: | Chylomicron remnants and very low density lipoprotein (VLDL) remnants are rapidly removed from the circulation by receptor-mediated endocytosis in the liver. Apolipoprotein E, a main apoprotein of the chylomicron, binds to a specific receptor on liver cells and peripheral cells. ApoE is essential for the normal catabolism of triglyceride-rich lipoprotein constituents. The APOE gene is mapped to chromosome 19 in a cluster with APOC1 and APOC2. Defects in apolipoprotein E result in familial dysbetalipoproteinemia, or type III hyperlipoproteinemia (HLP III), in which increased plasma cholesterol and triglycerides are the consequence of impaired clearance of chylomicron and VLDL remnants. [provided by RefSeq] |
| UniProt Code: | P02649 |
| NCBI GenInfo Identifier: | 114039 |
| NCBI Gene ID: | 348 |
| NCBI Accession: | P02649.1 |
| UniProt Secondary Accession: | P02649,Q9P2S4, B2RC15, C0JYY5, |
| UniProt Related Accession: | P02649,Q13791,Q6LA97,Q6LBZ1,Q8TCZ8 |
| Molecular Weight: | |
| NCBI Full Name: | Apolipoprotein E |
| NCBI Synonym Full Names: | apolipoprotein E |
| NCBI Official Symbol: | APOE |
| NCBI Official Synonym Symbols: | AD2; LPG; LDLCQ5; MGC1571 |
| NCBI Protein Information: | apolipoprotein E; apo-E; apolipoprotein E3; OTTHUMP00000159143; OTTHUMP00000197075; OTTHUMP00000197076; OTTHUMP00000197077 |
| UniProt Protein Name: | Apolipoprotein E |
| Protein Family: | Apolipoprotein |
| UniProt Gene Name: | APOE |
| UniProt Entry Name: | APOE_HUMAN |
| Component | Quantity (96 Assays) | Storage |
| ELISA Microplate (Dismountable) | 8×12 strips | -20°C |
| Lyophilized Standard | 2 | -20°C |
| Sample Diluent | 20ml | -20°C |
| Assay Diluent A | 10mL | -20°C |
| Assay Diluent B | 10mL | -20°C |
| Detection Reagent A | 120µL | -20°C |
| Detection Reagent B | 120µL | -20°C |
| Wash Buffer | 30mL | 4°C |
| Substrate | 10mL | 4°C |
| Stop Solution | 10mL | 4°C |
| Plate Sealer | 5 | - |
Other materials and equipment required:
- Microplate reader with 450 nm wavelength filter
- Multichannel Pipette, Pipette, microcentrifuge tubes and disposable pipette tips
- Incubator
- Deionized or distilled water
- Absorbent paper
- Buffer resevoir
*Note: The below protocol is a sample protocol. Protocols are specific to each batch/lot. For the correct instructions please follow the protocol included in your kit.
Allow all reagents to reach room temperature (Please do not dissolve the reagents at 37°C directly). All the reagents should be mixed thoroughly by gently swirling before pipetting. Avoid foaming. Keep appropriate numbers of strips for 1 experiment and remove extra strips from microtiter plate. Removed strips should be resealed and stored at -20°C until the kits expiry date. Prepare all reagents, working standards and samples as directed in the previous sections. Please predict the concentration before assaying. If values for these are not within the range of the standard curve, users must determine the optimal sample dilutions for their experiments. We recommend running all samples in duplicate.
| Step | |
| 1. | Add Sample: Add 100µL of Standard, Blank, or Sample per well. The blank well is added with Sample diluent. Solutions are added to the bottom of micro ELISA plate well, avoid inside wall touching and foaming as possible. Mix it gently. Cover the plate with sealer we provided. Incubate for 120 minutes at 37°C. |
| 2. | Remove the liquid from each well, don't wash. Add 100µL of Detection Reagent A working solution to each well. Cover with the Plate sealer. Gently tap the plate to ensure thorough mixing. Incubate for 1 hour at 37°C. Note: if Detection Reagent A appears cloudy warm to room temperature until solution is uniform. |
| 3. | Aspirate each well and wash, repeating the process three times. Wash by filling each well with Wash Buffer (approximately 400µL) (a squirt bottle, multi-channel pipette,manifold dispenser or automated washer are needed). Complete removal of liquid at each step is essential. After the last wash, completely remove remaining Wash Buffer by aspirating or decanting. Invert the plate and pat it against thick clean absorbent paper. |
| 4. | Add 100µL of Detection Reagent B working solution to each well. Cover with the Plate sealer. Incubate for 60 minutes at 37°C. |
| 5. | Repeat the wash process for five times as conducted in step 3. |
| 6. | Add 90µL of Substrate Solution to each well. Cover with a new Plate sealer and incubate for 10-20 minutes at 37°C. Protect the plate from light. The reaction time can be shortened or extended according to the actual color change, but this should not exceed more than 30 minutes. When apparent gradient appears in standard wells, user should terminatethe reaction. |
| 7. | Add 50µL of Stop Solution to each well. If color change does not appear uniform, gently tap the plate to ensure thorough mixing. |
| 8. | Determine the optical density (OD value) of each well at once, using a micro-plate reader set to 450 nm. User should open the micro-plate reader in advance, preheat the instrument, and set the testing parameters. |
| 9. | After experiment, store all reagents according to the specified storage temperature respectively until their expiry. |
When carrying out an ELISA assay it is important to prepare your samples in order to achieve the best possible results. Below we have a list of procedures for the preparation of samples for different sample types.
| Sample Type | Protocol |
| Serum | If using serum separator tubes, allow samples to clot for 30 minutes at room temperature. Centrifuge for 10 minutes at 1,000x g. Collect the serum fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. If serum separator tubes are not being used, allow samples to clot overnight at 2-8°C. Centrifuge for 10 minutes at 1,000x g. Remove serum and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. |
| Plasma | Collect plasma using EDTA or heparin as an anticoagulant. Centrifuge samples at 4°C for 15 mins at 1000 × g within 30 mins of collection. Collect the plasma fraction and assay promptly or aliquot and store the samples at -80°C. Avoid multiple freeze-thaw cycles. Note: Over haemolysed samples are not suitable for use with this kit. |
| Urine & Cerebrospinal Fluid | Collect the urine (mid-stream) in a sterile container, centrifuge for 20 mins at 2000-3000 rpm. Remove supernatant and assay immediately. If any precipitation is detected, repeat the centrifugation step. A similar protocol can be used for cerebrospinal fluid. |
| Cell culture supernatant | Collect the cell culture media by pipette, followed by centrifugation at 4°C for 20 mins at 1500 rpm. Collect the clear supernatant and assay immediately. |
| Cell lysates | Solubilize cells in lysis buffer and allow to sit on ice for 30 minutes. Centrifuge tubes at 14,000 x g for 5 minutes to remove insoluble material. Aliquot the supernatant into a new tube and discard the remaining whole cell extract. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Tissue homogenates | The preparation of tissue homogenates will vary depending upon tissue type. Rinse tissue with 1X PBS to remove excess blood & homogenize in 20ml of 1X PBS (including protease inhibitors) and store overnight at ≤ -20°C. Two freeze-thaw cycles are required to break the cell membranes. To further disrupt the cell membranes you can sonicate the samples. Centrifuge homogenates for 5 mins at 5000xg. Remove the supernatant and assay immediately or aliquot and store at -20°C or -80°C. |
| Tissue lysates | Rinse tissue with PBS, cut into 1-2 mm pieces, and homogenize with a tissue homogenizer in PBS. Add an equal volume of RIPA buffer containing protease inhibitors and lyse tissues at room temperature for 30 minutes with gentle agitation. Centrifuge to remove debris. Quantify total protein concentration using a total protein assay. Assay immediately or aliquot and store at ≤ -20 °C. |
| Breast Milk | Collect milk samples and centrifuge at 10,000 x g for 60 min at 4°C. Aliquot the supernatant and assay. For long term use, store samples at -80°C. Minimize freeze/thaw cycles. |